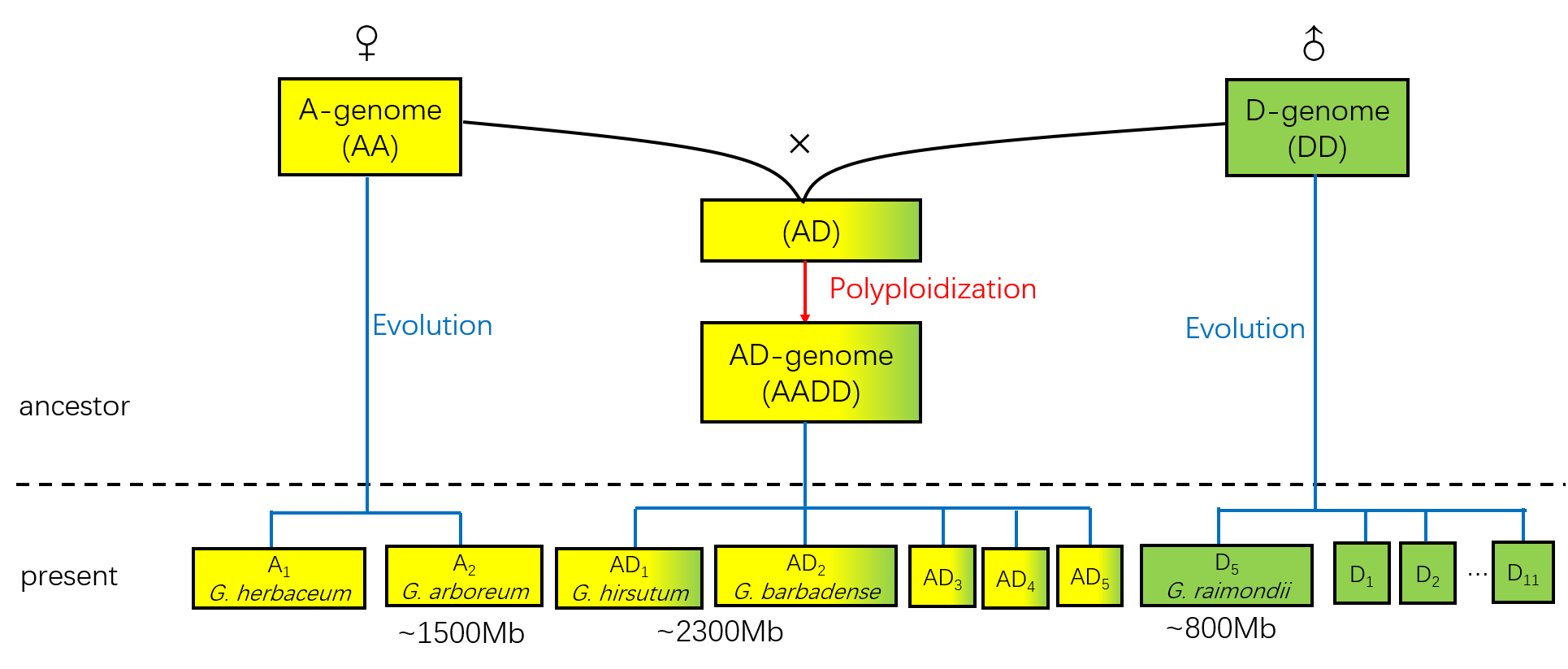
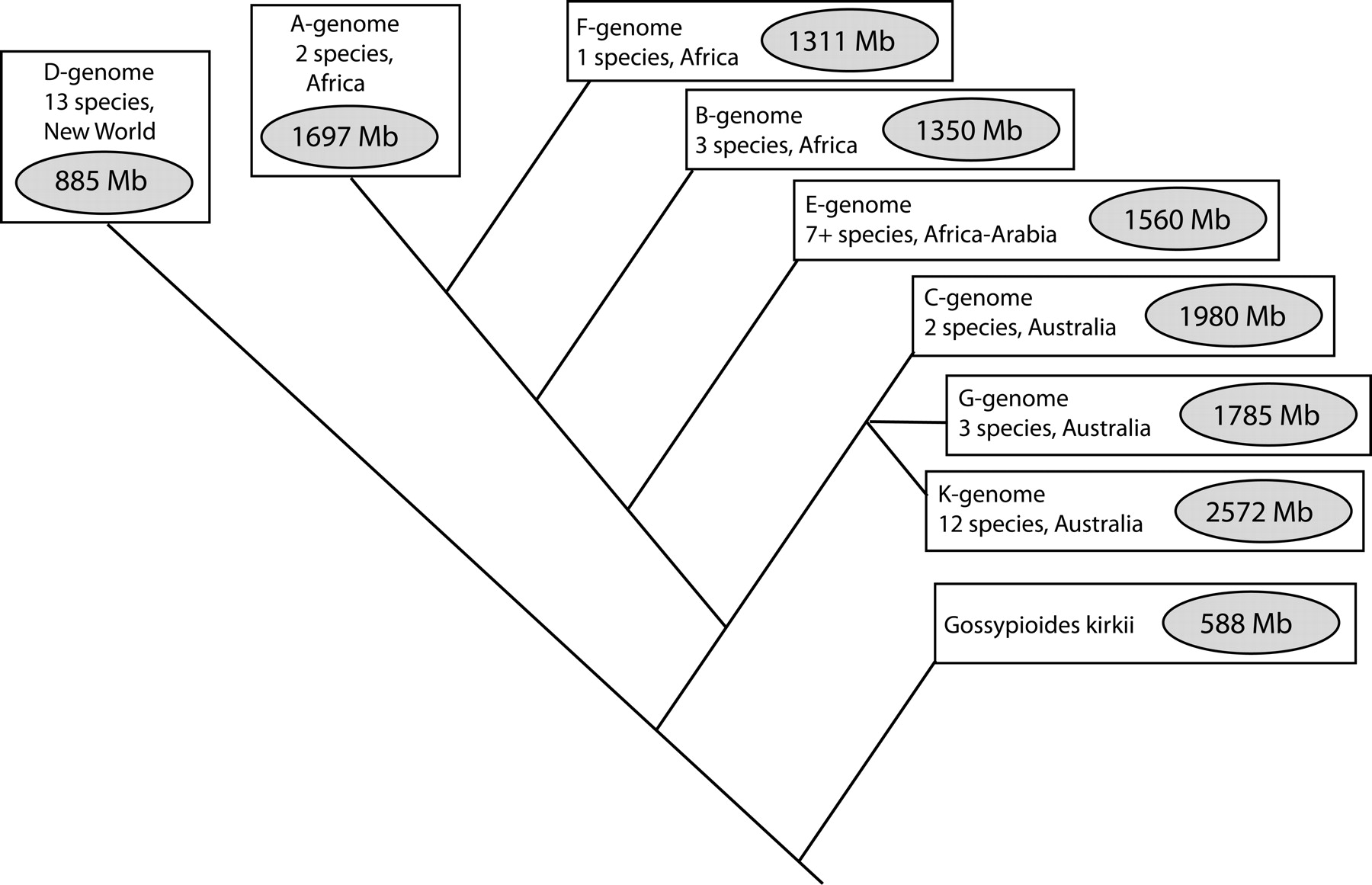
Cotton Species
Diploids: A2 and D5
The Gossypium genus contains about 50 speciesWendel J F, Albert V A. Phylogenetics of the cotton genus (Gossypium): character-state weighted parsimony analysis of chloroplast-DNA restriction site data and its systematic and biogeographic implications [J]. Syst Bot, 1992: 115-143.. Most of them are diploids and can be grouped to 8 diploid members (marked as A~G and K). They have different geographical origins and variable estimated genome sizes (Fig 1.)Hawkins J S, Kim H, Nason J D, et al. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium [J]. Genome Res, 2006, 16(10): 1252-1261., mostly due to different amount of DNA repeat elements. Despite all this, they share a common chromosome number (n = 13) and high levels of gene synteny.Rong J, Abbey C, Bowers J E, et al. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium) [J]. Genetics, 2004, 166(1): 389-417., indicating that they have a common ancestor (Fig 1.).
Tetraploids: AD1 and AD2
The tetraploid cotton species are thought to formed by an allopolyploidization event about 1~2 million years ago, which involved a D-genome species as the pollen-providing parent and an A-genome species as the maternal parentChen Z J, Scheffler B E, Dennis E, et al. Toward sequencing cotton (Gossypium) genomes [J]. Plant Physiol, 2007, 145(4): 1303-1310.(Fig 2.). Their descendants evolve into the current 5 tetraploid cotton species (Marked as AD1~AD5). Among them, the upland cotton G. hirsutum (AD1) and sea-island cotton G. barbadense (AD2) have become the main cultivated cotton species.
Please refer to CottonGen database to view all cotton species.
Genome Assembly
List of cotton genome assemblies.
| Chromosome Set | Species | Strain | Symbol in CottonFGD | Sequencing Technology | Available Date | Reference |
|---|---|---|---|---|---|---|
| A2 | Gossypium arboreum | SXY1 | Illumina HiSeq 2000 | 2014-05 | Li et. al., Genome sequence of the cultivated cotton Gossypium arboreum. Nature Genetics. 46, 567–572. 2014 | |
| A2 | Gossypium arboreum | SXY1 | CRI | PacBio and Hi-C | 2018-05 | Du et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nature genetics. 2018 May 07. |
| A2 | Gossypium arboreum | SXY1 | PacBio and Hi-C | 2020-04 | Huang, G. et al., Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nature Genetics. 2020. | |
| A2 | Gossypium arboreum | SXY1 | Oxford Nanopore | 2021-08 | Wang, M. et al. Comparative genome analyses highlight transposon-mediated genome expansion and the evolutionary architecture of 3D genomic folding in cotton. Mol Biol Evol. 2021 May 11:msab128. | |
| D5 | Gossypium raimondii | D5-3 | Illumina | 2012-01 | Wang et. al., The draft genome of a diploid cotton Gossypium raimondii. Nature Genetics. 44, 1098–1103. 2012 | |
| D5 | Gossypium raimondii | Ulbr. | JGI | Illumina, Sanger, Roche 454 | 2012-12 | Paterson AH et al., "Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres.", Nature, 2012 Dec 20;492(7429):423-7 |
| D5 | Gossypium raimondii | D5-4 | PacBio | 2019-09 | Udall J A, Long E, Hanson C, et al. De Novo Genome Sequence Assemblies of Gossypium raimondii and Gossypium turneri[J]. G3: Genes, Genomes, Genetics, 2019, 9(10): 3079-3085. | |
| D5 | Gossypium raimondii | D5-8 | Illumina HiSeq2500 | 2021-02 | Grover, et al. Insights into the evolution of the New World diploid cottons (Gossypium, subgenus Houzingenia) based on genome sequencing. Genome Biol Evol. 2019 Jan 1;11(1):53-71. | |
| D5 | Gossypium raimondii | D502 | Oxford Nanopore | 2021-08 | Wang, M. et al. Comparative genome analyses highlight transposon-mediated genome expansion and the evolutionary architecture of 3D genomic folding in cotton. Mol Biol Evol. 2021 May 11:msab128. | |
| A1 | Gossypium herbaceum | Mutema | PacBio and Hi-C | 2020-04 | Huang, G. et al., Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nature Genetics. 2020. | |
| D1 | Gossypium thurberi | D1-35 | Illumina | 2018-11 | Grover, et al. Insights into the evolution of the New World diploid cottons (Gossypium, subgenus Houzingenia) based on genome sequencing. Genome Biol Evol. 2019 Jan 1;11(1):53-71. | |
| D10 | Gossypium turneri | D10-3 | PacBio | 2019-09 | Udall J A, Long E, Hanson C, et al. De Novo Genome Sequence Assemblies of Gossypium raimondii and Gossypium turneri[J]. G3: Genes, Genomes, Genetics, 2019, 9(10): 3079-3085. | |
| G2 | Gossypium australe | PA1801 | PacBio | 2019-09 | Cai Y, Cai X, Wang Q, et al. Genome sequencing of the Australian wild diploid species Gossypium australe highlights disease resistance and delayed gland morphogenesis[J]. Plant biotechnology journal, 2019. | |
| AD2 | Gossypium barbadense | 3-79 | PacBio | 2019-08 | NA | |
| AD2 | Gossypium barbadense | 3-79 | HAU | Illumina, PacBio, BioNano, Hi-C | 2018-12 | Wang M, Tu L, Yuan D, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense[J]. Nature genetics, 2019, 51(2): 224-229. |
| AD2 | Gossypium barbadense | 3-79 | Illumina | 2015-12 | Yuan D, Tang Z, Wang M, et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres[J]. Scientific reports, 2015, 5: 17662. | |
| AD2 | Gossypium barbadense | Hai7124 | ZJU | Illumina, BioNano, Hi-C | 2019-03 | Hu Y, Chen J, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature genetics, 2019, 51(4): 739-748. |
| AD2 | Gossypium barbadense | Pima90 | P90HEBAU | Illumina, BioNano, Hi-C | 2021-08 | |
| AD5 | Gossypium darwinii | 1808015.09 | 2019-08 | NA | ||
| AD1 | Gossypium hirsutum | Tm-1 | Illumina | 2015-04 | Li et. al., Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution Nature Biotechnology. 33, 524–530. 2015 | |
| AD1 | Gossypium hirsutum | Tm-1 | HAU | Illumina, PacBio, BioNano, Hi-C | 2018-12 | Wang M, Tu L, Yuan D, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense[J]. Nature genetics, 2019, 51(2): 224-229. |
| AD1 | Gossypium hirsutum | Tm-1 | NAU | Illumina | 2015-04 | Zhang et. al., Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology. 33, 531–537. 2015 |
| AD1 | Gossypium hirsutum | Tm-1 | JGI | Illumina, PacBio | 2017-01 | NA |
| AD1 | Gossypium hirsutum | Tm-1 | ZJU | Illumina, BioNano, Hi-C | 2019-03 | Hu Y, Chen J, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature genetics, 2019, 51(4): 739-748. |
| AD1 | Gossypium hirsutum | Tm-1 | CRI | Illumina, PacBio, Hi-C | 2019-07 | Yang Z, Ge X, Yang Z, et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars[J]. Nature communications, 2019, 10(1): 1-13. |
| AD1 | Gossypium hirsutum | ZM24 | Illumina, PacBio, Hi-C | 2019-07 | Yang Z, Ge X, Yang Z, et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars[J]. Nature communications, 2019, 10(1): 1-13. | |
| AD1 | Gossypium hirsutum | NDM8 | NDM8HEBAU | Illumina, PacBio, Hi-C | 2021-08 | |
| AD4 | Gossypium mustelinum | 1408120.09 | 2019-09 | NA | ||
| AD3 | Gossypium tomentosum | 7179.01 | 2020-01 | NA |
Gene Models
Gene ID
Due to the limit of data providers, currently CottonFGD only includes protein coding genes.
All gene IDs are directly imported from data provider annotations. Therefore, you can directly use these IDs to search in other cotton databases (e.g., CottonGen). Their formats are listed as follows:
| Species | Gene ID Format | Gene ID Example |
|---|---|---|
| G. hirsutum, CRI assembly | Gh_[%3c]G[%04d]00 |
Gh_A01G001100 |
| G. hirsutum, HAU assembly | Ghir_[%3c]G[%05d]0 |
Ghir_A01G000120 |
| G. hirsutum, JGI assembly | Gohir.[%03c]G[%06d] |
Gohir.A01G001300 |
| G. hirsutum, NAU assembly | Gh_[%3c]G[%04d] |
Gh_A01G0001 |
| G. hirsutum, ZJU assembly | GH_[%3c]G[%04d] |
GH_D07G0123 |
| G. hirsutum, NDM8HEBAU assembly | GhM_[%3c]G[%04d] |
GhM_D12G0324 |
| G. barbadense, HAU assembly | Gbar_[%3c]G[%05d]0 |
Gbar_A01G014970 |
| G. barbadense, ZJU assembly | GB_[%3c]G[%04d] |
GB_A01G0011 |
| G. barbadense, P30HEBAU assembly | GbM_[%3c]G[%04d] |
GbM_D13G0404 |
| G. arboreum, CRI assembly | Ga[%2c]G[%04d] |
Ga01G0012 |
| G. raimondii, JGI assembly | Gorai.[%03c]G[%06d] |
Gorai.001G000100 |
Transcript ID
IDs of transcripts are marked as .1, .2, .3,
..., appending to their gene IDs. IDs with .1 are usually the longest ones among all the isoforms (i.e., principle transcripts).
In order to maintain the consistence between the four cotton species, only principle transcripts are analyzed in CottonFGD.
Gene Name and Description
Each gene's name and description are based on its best homolog with Swiss-Prot proteins, as it is non-redundant and manually-reviewed. The homology is identified by NCBI BLAST+Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications [J]. BMC Bioinformatics, 2009, 10: 421.:
$ blastp -query [pep.fa] -db swissprotdb.fa -evalue 1e-5 -max_target_seqs 1 -max_hsps 1
For each Swiss-Prot entry, the "Gene names" value is served as cotton gene's name (symbol) while the "Protein names" value is served as cotton gene's description. All genes with Swiss-Prot hits must have gene descriptions, but some of them have no gene names. For example, gene Gh_A01G0010 in G. hirsutum has its best Swiss-Prot homolog P48504. It has the description (Protein names) Cytochrome b-c1 complex subunit 6, but it has no gene names.
Transcript Structure
All the transcript structures: exons, introns, coding regions and untranslated regions (UTRs) are extracted from data providers annotations. It should be noticed that limited by data provider, UTR annotations might not completed.
Gene Function Annotation
As mentioned above, only the principle transcript of each gene is analyzed. Thus, all gene function annotations such as homology, GO(Gene Ontology), InterPro and pathway are all based on the protein sequences of their principle transcripts.
Protein Property Statistics
CottonFGD includes the following protein property statistics for each gene:
- Residue Composition:
- The percentage of basic residues: His(H), Lys(K), Arg(R)
- The percentage of acidic residues: Asp(D), Glu(E)
- Molecular Weight (kDa)
- Charge
- Isoelectric Point: the pH value at which this molecule carries no net electrical charge
- Grand Average of Hydropathy: The sum of hydropathy values of all amino acids divided by the protein length. Positive value indicates hydrophobic.
The value of residue composition, molecular weight, charge and isoelectric point are calculated using pepstats in EMBOSS package (v6.5.7.0):
$ pepstats -sequence [pep.fa] -outfile [pep.out]
You can see an example of the output in the pepstats manual.
The value of Grand Average of Hydropathy is calculated using BioPerl (v1.6.924):
use Bio::SeqIO;
use Bio::Tools::SeqStats; # Package to calculate hydropathicity
my $seqio_obj = Bio::SeqIO->new(-file=>shift, -format=>"fasta");
while (my $seq_obj = $seqio_obj->next_seq) {
my $id = $seq_obj->display_id;
my $seq_stats = Bio::Tools::SeqStats->new(-seq => $seq_obj);
my $gravy;
eval{$gravy = $seq_stats->hydropathicity();};
}
Protein Domain, Gene Ontology & InterPro Items
The possible domain regions for each protein and the associated GO (Gene Ontology) / InterPro items are predicted using a locally installed copy of InterProScan (v5.16-55.0):
./interproscan.sh -dp -f tsv -goterms -i [pep.fa]
Currently InterProScan includes 15 types of domain databases:
| Database | Description | Accession ID Format | Accession ID Example |
|---|---|---|---|
| Coils (2.2.1) | Prediction of Coiled Coil Regions in Proteins | Coil | |
| Gene3D (3.5.0) | Structural assignment for whole genes and genomes using the CATH domain structure database | G3DSA:[%d.%d....] |
G3DSA:3.10.330.20 |
| Hamap (201511.02) | High-quality Automated and Manual Annotation of Microbial Proteomes | MF_[%05d] |
MF_01007 |
| PANTHER (10.0) | The PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System is a unique resource that classifies genes by their functions, using published scientific experimental evidence and evolutionary relationships to predict function even in the absence of direct experimental evidence. | PTHR[%5d]PTHR[%5d]:SF[%d]
|
PTHR12133 PTHR24279:SF100 |
| Pfam (28.0) | A large collection of protein families, each represented by multiple sequence alignments and hidden Markov models (HMMs) | PF[%05d] |
PF03061 |
| Phobius (1.01) | A combined transmembrane topology and signal peptide predictor | CYTOPLASMIC_DOMAIN NON_CYTOPLASMIC_DOMAIN SIGNAL_PEPTIDE_C_REGION SIGNAL_PEPTIDE_H_REGION SIGNAL_PEPTIDE_N_REGION SIGNAL_PEPTIDE TRANSMEMBRANE |
|
| PIRSF (3.01) | The PIRSF concept is being used as a guiding principle to provide comprehensive and non-overlapping clustering of UniProtKB sequences into a hierarchical order to reflect their evolutionary relationships. | PIRSF[%06d] |
PIRSF023803 |
| PRINTS (42.0) | A fingerprint is a group of conserved motifs used to characterise a protein family | PR[%05d] |
PR00109 |
| ProDom (2006.1) | ProDom is a comprehensive set of protein domain families automatically generated from the UniProt Knowledge Database. | PD[%06d] |
PD005155 |
| ProSite (20.113) | PROSITE consists of documentation entries describing protein domains, families and functional sites as well as associated patterns and profiles to identify them | PS[%05d] |
PS00036 |
| SignalP (4.1) | SignalP predicts the presence and location of signal peptide cleavage sites in amino acid sequences for eukaryotes, gram-positive prokaryotes or gram-negative prokaryotes | SignalP-noTM SignalP-TM |
|
| SMART (6.2) | SMART allows the identification and analysis of domain architectures based on Hidden Markov Models or HMMs | SM[%05d] |
SM00338 |
| SUPERFAMILY (1.75) | SUPERFAMILY is a database of structural and functional annotation for all proteins and genomes | SSF[%d] |
SSF53335 |
| TIGRFAM (15.0) | TIGRFAMs are protein families based on Hidden Markov Models or HMMs | TIGR[%05d] |
TIGR00006 |
| TMHMM (2.0c) | Prediction of transmembrane helices in proteins | Tmhelix |
The accession ID formats of GO (Gene Ontology) and InterPro items are listed as follows:
| Database | Accession ID Format | Accession ID Example |
|---|---|---|
| Gene Ontology | GO:[%07d] |
GO:0006629 |
| InterPro | IPR[%06d] |
IPR004299 |
Homology
Currently CottonFGD includes homology information for 22 other representative plant species from all the main categories: Eudicots, Monocots, Acrogymnospermae, Lycopodiidae, Bryophyta and Chlorophyta. Homology information are only available for genes in G. hirsutum, CRI assembly.
The best homolog for each cotton gene is searched using NCBI BLAST+, similar with defining gene names.
KEGG Pathway
The associated KEGG pathways for each gene is defined by two steps: First, all cotton genes are assigned
to KEGG Orthology using the
KEGG Automatic Annotation Server. We select all the
available plant species as our "GENES data set". For each gene, only one KEGG Orthology item
(ID Format: K[%05d]) is assigned. Then, the KEGG Orthology item is mapped to its associated
KEGG Pathways (ID Format: map[%05d] or
ko[%05d]) and KEGG Modules (ID Format: M[%05d])
Gene Expression
The expression patterns for each gene (i.e. principle transcript) are measured using RNA-seq datasets from SRP166405 Hu Y, Chen J, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature genetics, 2019, 51(4): 739-748.. This dataset contains 49 samples each with 1-3 biological replicates. Expression data are only available for genes in G. hirsutum, CRI assembly.
| Type | Sample | Num. of biological Replicates | SRA IDs |
|---|---|---|---|
| Tissue and Organ | anther | 3 | SRR8089834; SRR8089835; SRR8089836 |
| Tissue and Organ | bract | 3 | SRR8089862; SRR8089861; SRR8089833 |
| Tissue and Organ | filament | 3 | SRR8089837; SRR8089838; SRR8089839 |
| Tissue and Organ | leaf | 3 | SRR8089855; SRR8090014; SRR8089903 |
| Tissue and Organ | pental | 3 | SRR8089902; SRR8089868; SRR8089867 |
| Tissue and Organ | pistil | 1 | SRR8089840 |
| Tissue and Organ | root | 3 | SRR8089897; SRR8089896; SRR8089895 |
| Tissue and Organ | sepal | 3 | SRR8089863; SRR8089866; SRR8089865 |
| Tissue and Organ | stem | 3 | SRR8089977; SRR8089975; SRR8089969 |
| Tissue and Organ | torus | 3 | SRR8089870; SRR8089869; SRR8089864 |
| Fiber development | fiber at 10DPA | 3 | SRR8090044; SRR8090041; SRR8090042 |
| Fiber development | fiber at 15DPA | 3 | SRR8090046; SRR8090049; SRR8090050 |
| Fiber development | fiber at 20DPA | 3 | SRR8090004; SRR8090007; SRR8090006 |
| Fiber development | fiber at 25DPA | 1 | SRR8090010 |
| Drought stress | leave under drought stress for 1h | 3 | SRR8089985; SRR8089984; SRR8089987 |
| Drought stress | leave under drought stress for 3h | 3 | SRR8089986; SRR8089989; SRR8089988 |
| Drought stress | leave under drought stress for 6h | 3 | SRR8089991; SRR8089990; SRR8089983 |
| Drought stress | leave under drought stress for 12h | 3 | SRR8089982; SRR8090019; SRR8090020 |
| Drought stress | leave under drought stress for 24h | 3 | SRR8090021; SRR8090022; SRR8090015 |
| Cold stress | leaves cold-treated for 1h | 3 | SRR8089823; SRR8089824; SRR8089825 |
| Cold stress | leaves cold-treated for 3h | 3 | SRR8089826; SRR8089827; SRR8089828 |
| Cold stress | leaves cold-treated for 6h | 3 | SRR8089829; SRR8089830; SRR8089831 |
| Cold stress | leaves cold-treated for 12h | 3 | SRR8089832; SRR8089924; SRR8089923 |
| Cold stress | leaves cold-treated for 24h | 3 | SRR8089922; SRR8089921; SRR8089920 |
| control for stress | leaves control 0h | 3 | SRR8090035; SRR8090032; SRR8090033 |
| control for stress | leaves control 1h | 3 | SRR8090030; SRR8090031; SRR8090039 |
| control for stress | leaves control 3h | 3 | SRR8090040; SRR8090074; SRR8090073 |
| control for stress | leaves control 6h | 3 | SRR8090076; SRR8090075; SRR8090070 |
| control for stress | leaves control 12h | 3 | SRR8090069; SRR8090072; SRR8090071 |
| control for stress | leaves control 24h | 2 | SRR8090078; SRR8090077 |
| Heat stress | leaves heat-treated for 1h | 3 | SRR8089919; SRR8089918; SRR8089917 |
| Heat stress | leaves heat-treated for 3h | 3 | SRR8089916; SRR8089915; SRR8089953 |
| Heat stress | leaves heat-treated for 6h | 3 | SRR8089954; SRR8089951; SRR8089952 |
| Heat stress | leaves heat-treated for 12h | 3 | SRR8089957; SRR8089958; SRR8089955 |
| Heat stress | leaves heat-treated for 24h | 3 | SRR8089956; SRR8089949; SRR8089950 |
| salt stress | leaves salt-treated for 1h | 3 | SRR8090016; SRR8090017; SRR8090018 |
| salt stress | leaves salt-treated for 3h | 3 | SRR8090026; SRR8090027; SRR8090056 |
| salt stress | leaves salt-treated for 6h | 3 | SRR8090055; SRR8090054; SRR8090053 |
| salt stress | leaves salt-treated for 12h | 2 | SRR8090060; SRR8090059 |
| salt stress | leaves salt-treated for 24h | 3 | SRR8090058; SRR8090057; SRR8090064 |
| ovule development | ovule at -3DPA | 2 | SRR8089841; SRR8089842 |
| ovule development | ovule at 0DPA | 3 | SRR8090087; SRR8090086; SRR8090085 |
| ovule development | ovule at 1DPA | 3 | SRR8090084; SRR8090083; SRR8090082 |
| ovule development | ovule at 3DPA | 2 | SRR8090081; SRR8090080 |
| ovule development | ovule at 5DPA | 3 | SRR8090089; SRR8090088; SRR8090043 |
| ovule development | ovule at 10DPA | 3 | SRR8090047; SRR8090048; SRR8090045 |
| ovule development | ovule at 15DPA | 3 | SRR8090003; SRR8090002; SRR8090005 |
| ovule development | ovule at 20DPA | 3 | SRR8090009; SRR8090008; SRR8090011 |
| ovule development | ovule at 25DPA | 1 | SRR8089973 |
TPM values are calculated using salmon Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., & Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods. (v1.1.0).
salmon quant -i cDNA.Ghir.CRI -l A -1 reads.1.fq.gz -2 reads.2.fq.gz --seqBias --gcBias --validateMappings -o output
The generated files quant.sf are available in the Download page.
Web Browser Compatibility
CottonFGD is designed to be compatible with all modern web browsers (such as Mozilla Firefox, Google Chrome, Sarifi, Microsoft Edge, etc ...) on a variety of devices (such as PC, tablet and mobile). Except for some subtle differences on front-end appearance, the recent version of Microsoft Internet Explorer (later than version 9.0) is also acceptable. It is strongly not recommended to use old versions of browsers (such as Internet Explorer lower than 8.0) as this would encounter many unnecessary bugs.
Tips
- "JavaScript" must be turned on, otherwise you can not do anything. By default it is turned on at almost all browsers.
- "Cookie" is recommended to be turned on. It is used to "remember" your previous settings on several tools. By default it is turned on at almost all browsers.