Input
- Users should input a list of cotton gene/transcript IDs by typing/paste to the textarea or upload a text file, one ID per line.
- If both textarea input and file upload are done, CottonFGD would combine the IDs.
- Flanking blanks and empty lines are automatically removed. Duplicated IDs are cleaned.
- In Data Fetch Option, ≤ 1000 IDs are allowed. IDs from different species/assemblies are also acceptable.
- In Enrichment Analysis, the number of IDs is not limited. However they must come from the same species and assembly.
Fetch Information
In this section, users should specify the fetch options by clicking the radios and/or setting possible parameters. Currently CottonFGD supports fetching information for the following categories:
- Gene/Transcript Features:
- Gene Feature: Name, Description, Species, Genomic Locations, Gene Length
- Transcript Feature: Transcript/CDS Length, GC Content, Exon/Intron Number, Mean Exon/Intron Length
- Protein Statistics: Protein Length, Molecular Weight, Charge, Hydropathy, Isoelectric Point
- Sequences: Genomic DNA, Transcript, CDS, Protein, 5' and 3'UTR, Intron, Upstream/Downstream (Customized length)
- Domain
- Homology
- Function: GO, InterPro and pathway
- Expression
Except for fetching sequence, all the other fetching options would return a table that list fetched results in the same order as input IDs. For genes with not available information, this row would show as "NA".
In the sequence fetch section, results would be returned in FASTA format text.
Enrichment Analysis
In this section, CottonFGD conducts the enrichment analysis based on users' input IDs and their selected background genome.
The P value for each functional item (GO, InterPro or Pathway) is calculated based on its proportion in the query and background genes
using the phyper function in R and adjusted by p.adjust function.
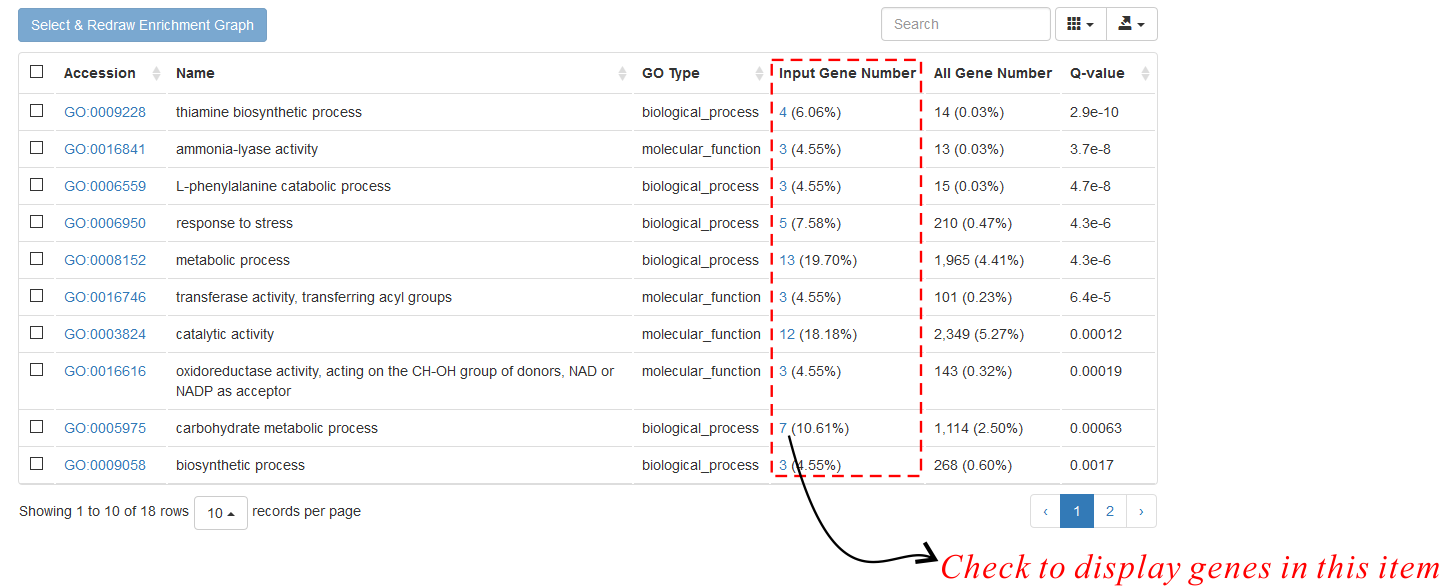
The returned results contain a table listing significantly enriched items and a column chart. In the table (Fig. 1),
there is a column showing the number of query genes in each function item. Users can click the attached hyperlinks to see the detailed
gene list. Note: the minimum value (> 3) of this column is controlled by the parameter Minimum Gene Number for each Analyzed Term.
It could be customized, but must > 3, as too few genes is not meaningful in statistics.