Gene Name/Description
Each gene's name and description are based on its best homolog with Swiss-Prot proteins . Generally speaking, names are short symbols (such as ACT2), while descriptions are full words (such as Actin-2).
After users submit their query names/descriptions, CottonFGD would return a list of genes that match inputted names.
Domain
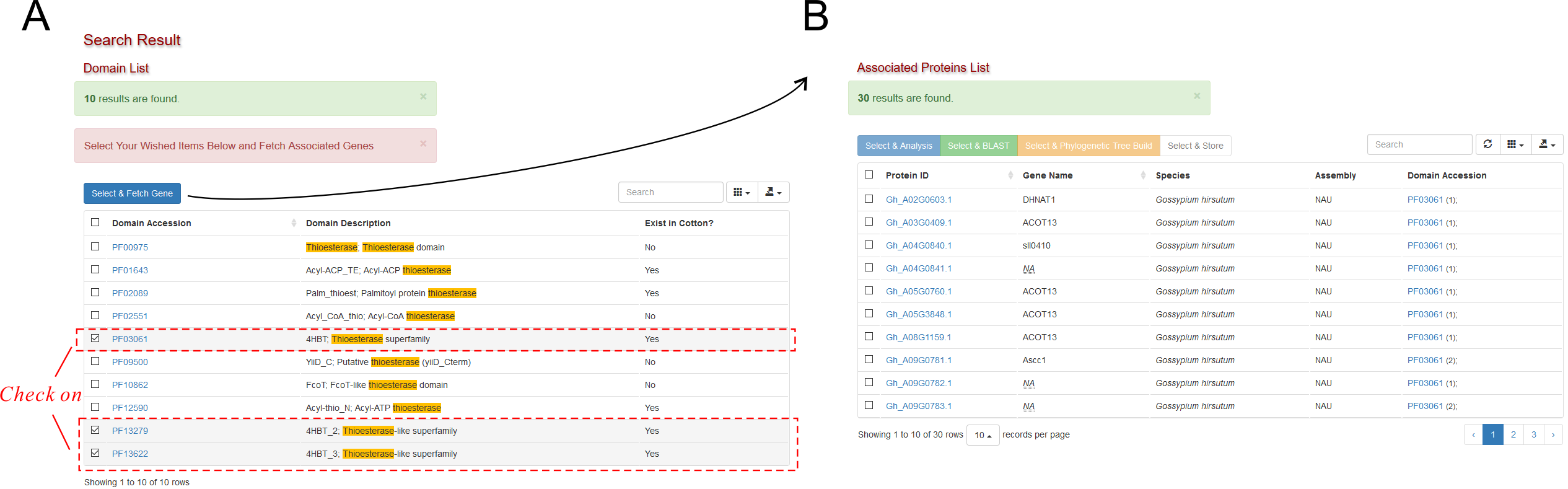
CottonFGD supports searching domains from 15 different kind of databases .
Two-step Search
Searching by domain ID is more preferred. If you want to search by domain name, you might need conduct a two-step search:
Gene Ontology, InterPro and Pathway Item
Gene Ontology and InterPro items are annotated by InterProScan . KEGG pathway items are annotated by KAAS . Similar as the domain search, two-step search is also used here.
Expression Profiles
Gene expression data are from 168 RNA-seq runs in 20 experiment groups, covering all the 4 cotton species. .
By Expression Levels
In this section, users can search genes that are expressed in certain experiment conditions (Tissue/Organ, development stage, Stress treatment) by a customized threshold (default threshold is FPKM ≥ 1). Multiple selection means expressed in all the selected experiments.
By Expression Difference
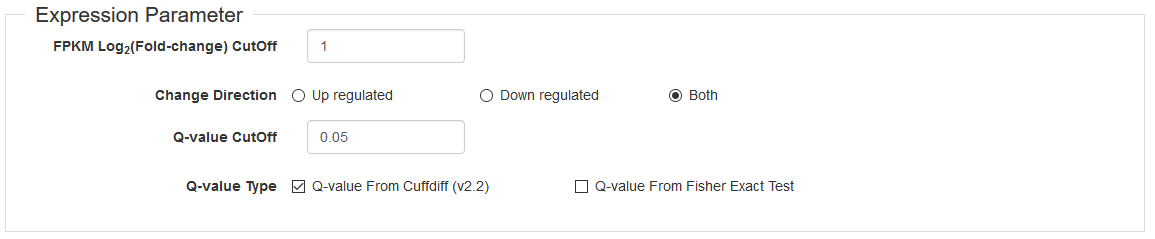
In this section, users can search genes that are significantly up/down-regulated by several customized thresholds (Fig. 2) in a pair of selected experiments. These parameters include:
- FPKM Log2(Fold-change): Default is ≥ 1
- Direction: Up and/or down regulated
- Q-value: Default is < 0.05
- Q-value type: Whether use Q-value from Cufflinks' output and/or from Fisher's Exact Test. Default is use Cufflinks' output only. Fisher's Exact Test should only be used if no results are found from Cufflinks' output. See the explaination.
Multiple Selection is only Allowed within the same Experiment Group
In order to reduce errors introduced among different sequencing strategy and depth, multiple selection is only allowed within the same experiment group.
In addition, for time-series experiment group, you can only see expression differences between the adjacent time points.