The web browser would remember your last settings
According to the JBrowse's default design, the web browser would remember your last settings, such as track on/off and fold/unfold.
Overview
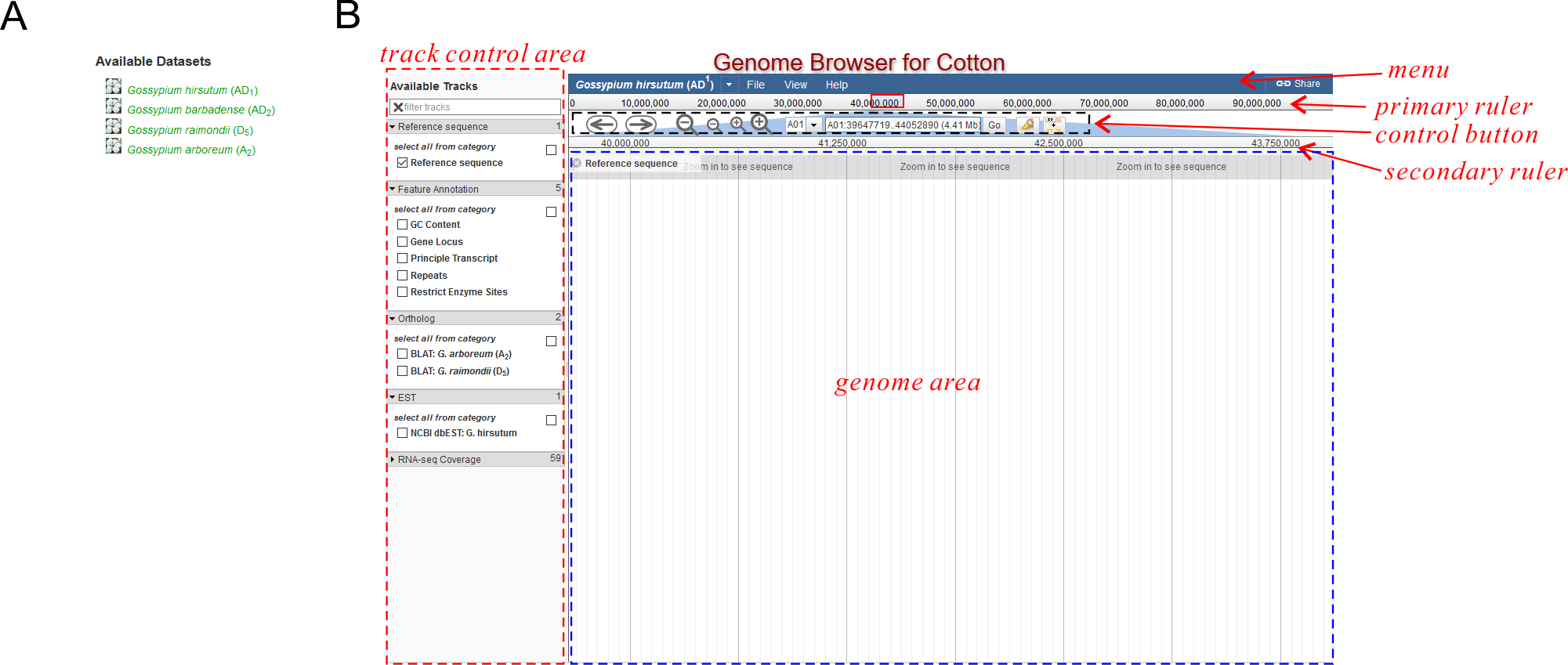
JBrowse is based on an reference genome of a certain species. Therefore when visiting the hompage, you can only see a species list (See Fig. 1A) instead of the real browser. Choose what you want and then you will see the main browser (Fig. 1B) for this species.
Tracks
JBrowse stores all kinds of data (reference sequence, gene locus, transcript structure, gene expression, etc) on a variety of Tracks. You can turn on/off any tracks using the left-side track control area (See Fig. 1B), and the relevant tracks would appear/disappear on the genome area. When you first open JBrowse, you might only see the "Reference Sequence" track appeared. After that, JBrowse would "remember" your last opened/closed tracks by using cookies on your browsers.
Track Control Area
In the track control area, tracks are grouped to several categories, which could be collapsed/expanded by clicking the arrows (Fig. 2). Some categories contain further sub-categories, making a so-called "hierarchical structure". For example, By default, all tracks options are expanded except for the "RNA-seq Coverage" category which includes huge number of samples. You can collapse/expand any categories you want, and JBrowse would "remember" your last operation by using cookies on your browsers.
Track Label and Menu
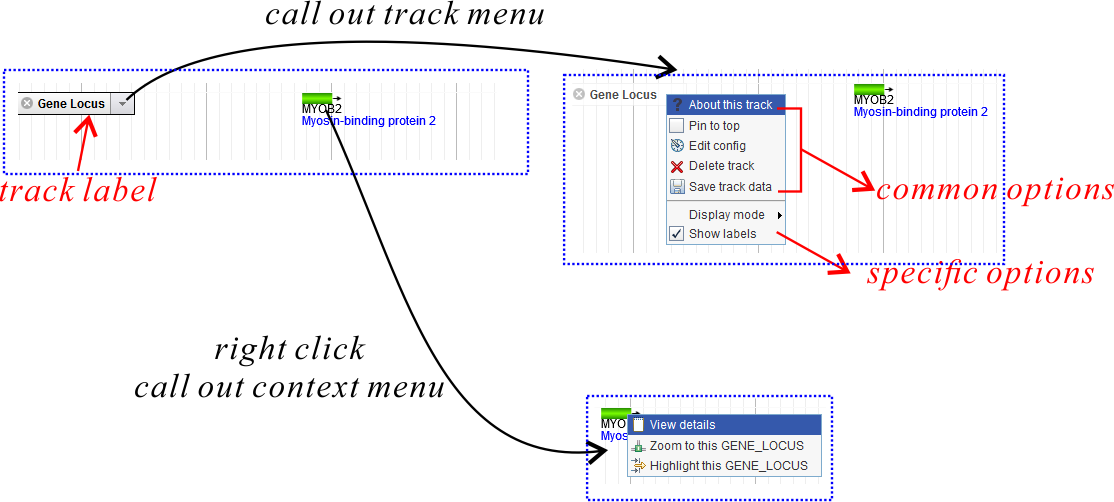
When you turn on a track (for example, Gene Locus), it would appear on the genome area with a track label
(Fig. 3). Clicking the "Close" button  on
label would turn off this track. If the track name is very long, the track label might overlap with the
rendered track images. In this situation you can use the "Hide/Show Track Titles" button to
hide all track labels.
on
label would turn off this track. If the track name is very long, the track label might overlap with the
rendered track images. In this situation you can use the "Hide/Show Track Titles" button to
hide all track labels.
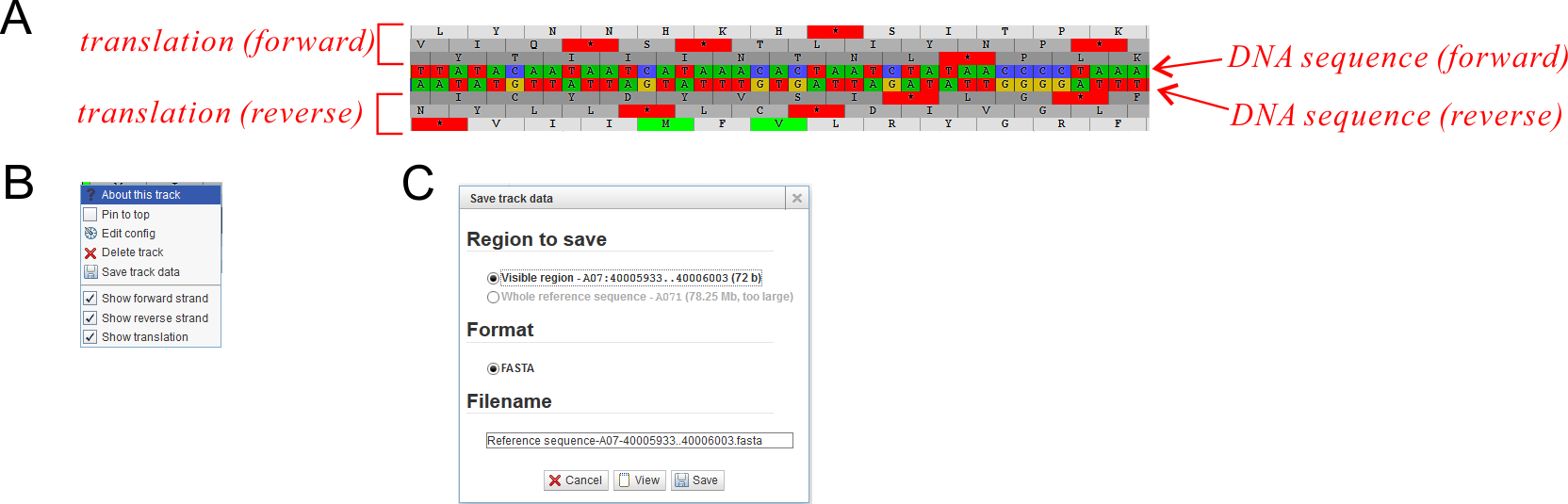
Clicking the arrow on the track label would call out the track label menu (Fig. 3). Besides five common menu options existing in all track menus, there might also be several options restricted in certain tracks. Some options are selectable or even editable, but only valid for the current session. The five common menu options are:
- About this track: Show some basic information (and sometimes including statistics information) for this track.
- Pin to top: Keep this track on top of all other tracks. By default tracks are added and appeared in turn according to users' operation on the Track Control Area.
- Edit config: For end users, don't try to edit it. Otherwise you should referred to the configuration manual carefully.
- Delete track: Similar as to close the track, however it would be deleted from the Track Control Area and you can not turn it on again unless refresh the web page.
- Save track data: Export track data of the current region or the whole reference region to an external text file. The data format is changed according to different track types.
In some tracks (mainly the FeatureBox track), right-click on the rendered item would call out a context menu (Fig. 3). See FeatureBox track for more details.
Track Type
Currently JBrowse in CottonFGD includes three track types:
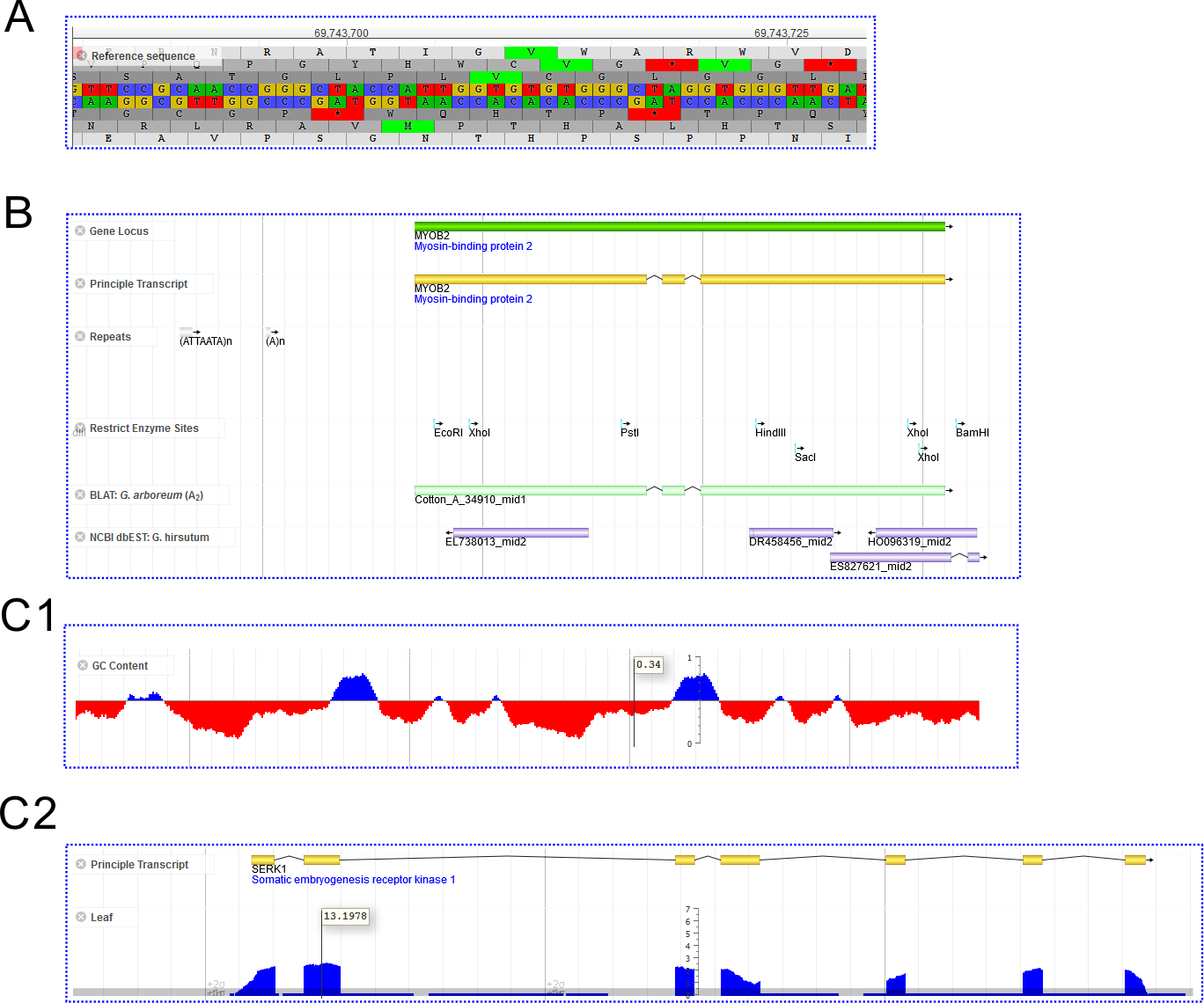
- Reference Sequence (Fig. 4A): This track displays the double strand reference sequences and the six-frame translation protein sequences. Usually only one reference sequence track in a JBrowse instance.
- FeatureBox (Fig. 4B): These tracks display feature regions (for example, genes, transcripts, repeat elements, etc) on the genome, rendering as colored boxes with arrows indicating forward or reverse strands.
- XYPlot (Fig. 4C1,2): These tracks display quantitative data (Y axis) distribution among the reference genome (X axis). Hovering on it would show y values
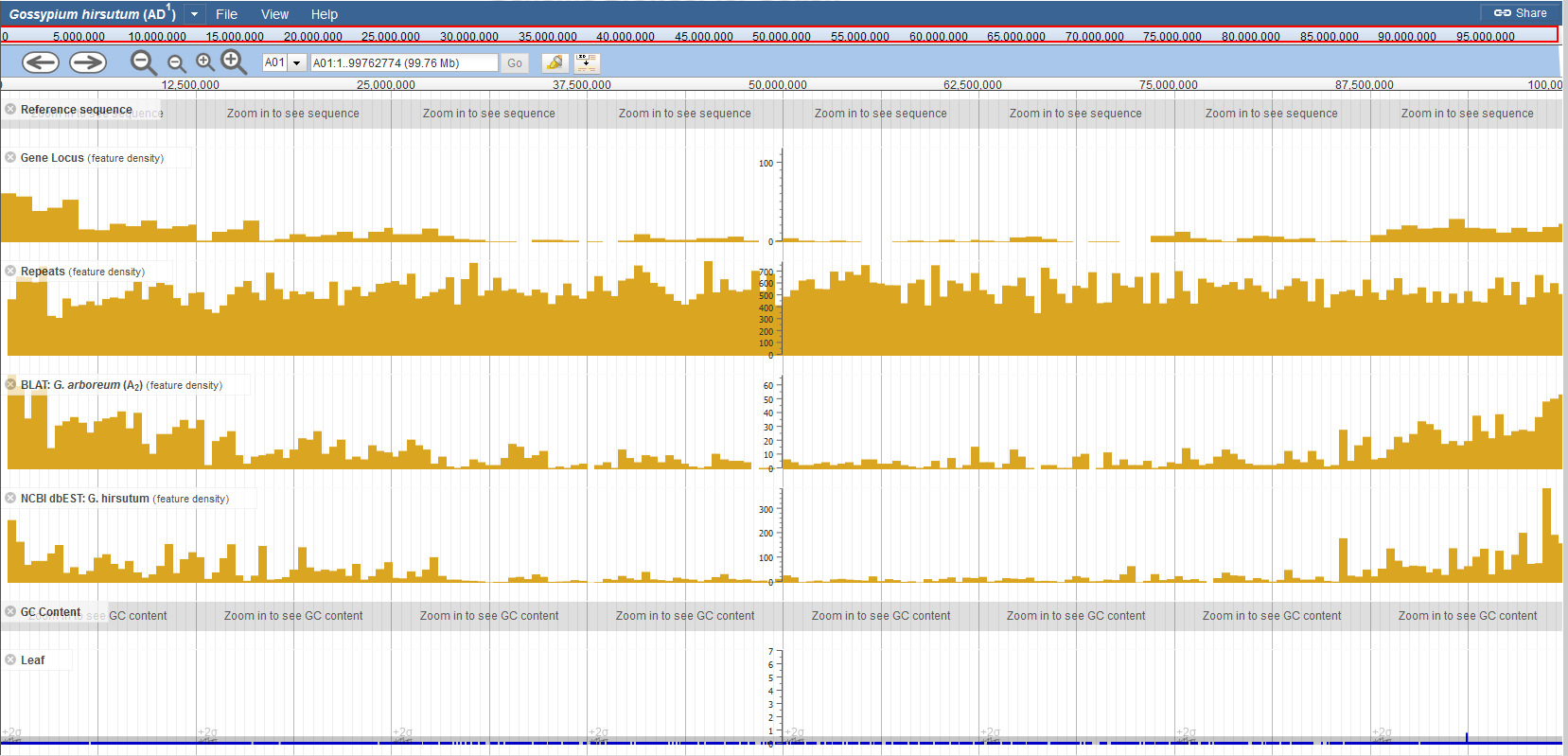
If you are viewing on a large region (for example, change to a new chromosome, or zoom out to the highest level), most tracks might show histograms indicating the feature density, instead of filling large numbers of features on the whole screen (Fig. 5).
Menu
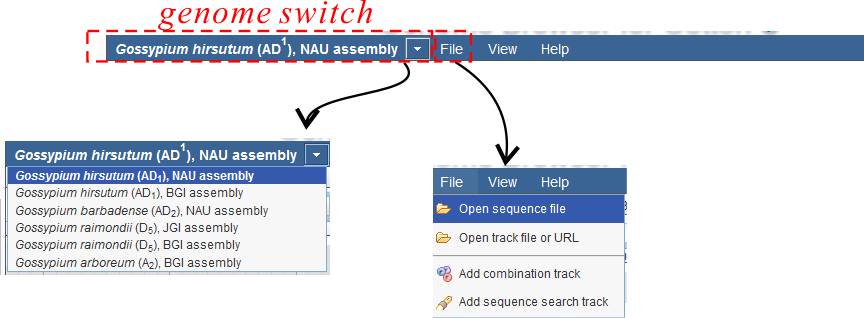
The menu section (Fig. 6) mainly consists of:
- Genome Switch Section: Here you can switch between different genomes
- Menu: File
- Open sequence file: Here you can upload your own sequence file. (Do not recommend if your file is very large)
- Add sequence search track: Here you can search short sequence segments in both forward and reverse directions
- Other Menu: such as Help
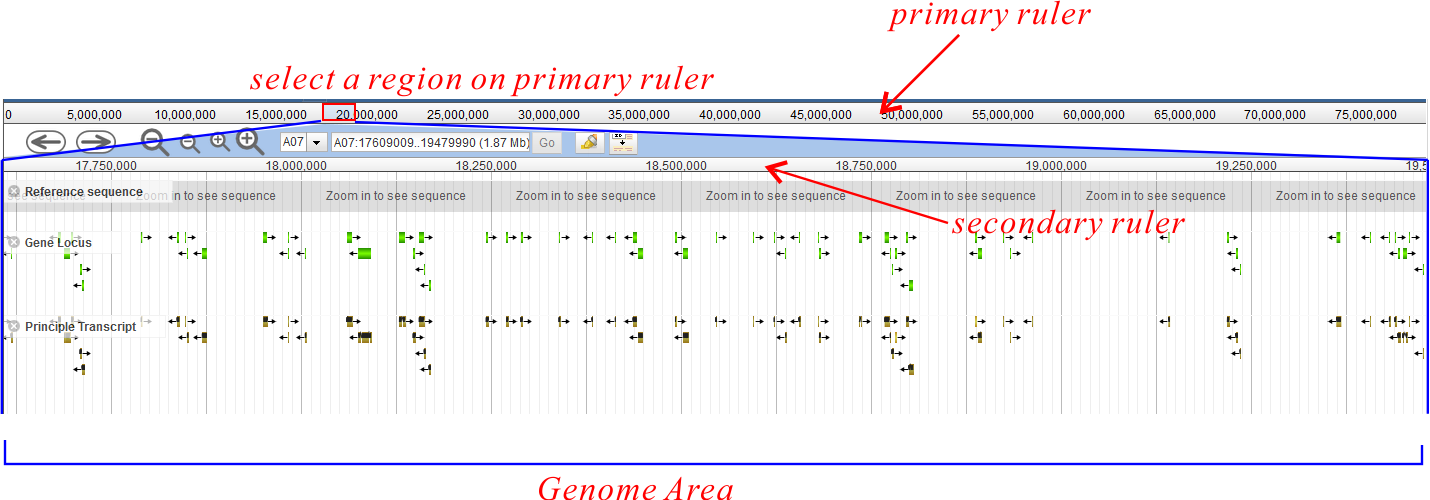
Ruler
There are two rulers (Fig. 1): a primary ruler and a secondary ruler. Between them is a set of control button.
The primary ruler extends to the full length of the current chromosome. Users can drag to select a sub-region on it and the secondary ruler displays the selected sub-region (Fig. 7). The genome area only displays features within the secondary ruler region.
Action
Possible action buttons are:
- Moving
 : Use them to move in the genome (It is also acceptable to drag on the genome area or select on the
ruler directly.
: Use them to move in the genome (It is also acceptable to drag on the genome area or select on the
ruler directly. - Zoming
 : Use them to zoom in/out
: Use them to zoom in/out - Navigation Bar: Input genomic positions in the following formats:
A04: Jump to chromosome A04.A04:1000000..2000000: Jump to the region on chromosome A04 between 1Mb and 2MbGh_A01G0001: Jump to gene Gh_A01G0001 (Gene ID, transcript ID is also OK)ACT4: Jump to gene ACT4 (Gene Name, might relect multiple positions)
- Highlight
 : Use it to highlight a region. Click it first and then
select a region in the genome area using mouse. (However, you can also highlight a region using the View Menu )
: Use it to highlight a region. Click it first and then
select a region in the genome area using mouse. (However, you can also highlight a region using the View Menu ) - Hide/Show Track Label
 : Using this button to hide/show
track labels
: Using this button to hide/show
track labels
Reference Sequence
This track shows reference sequence (DNA) in both forward and reverse strand and in six-frame translations (Fig. 8A Only visible when zoom in to a short region length). Display options could be customized by the track menu (Fig. 8B).
Save Track Data
In this track, data would be saved as FASTA format sequence of the current region (Fig. 8C).
Track Type: FeatureBox
This track displays qualitative features in a genome region (for example, genes, repeat elements, ...). It renders as long boxes in colors. These tracks include:
- Gene Locus;
- Principle Transcript; (And also other non-principle transcripts in G. raimondii JGI)
- Repeat Elements
- Restrict Enzyme Sites (G. hirsutum NAU only): annotated for seven enzymes using the
restrictprogram in EMBOSS package:restrict -enzymes EcoRI,HindIII,PstI,XhoI,BamHI,SacI,AvaIII -sitelen 4 -noambiguity
- Ortholog (G. hirsutum NAU only): Mapping the CDS sequences of A2 (G. arboreum) and D5 (G. raimondii) to AD1(G. hirsutum) using BLAT
- Cufflinks Merge Transcripts From RNA-seq: These are transcripts from cufflinks merge result:
cufflinks --GTF-guide=[gene.gff] --multi-read-correct --frag-bias-correct=[refer.genome.fa] --min-intron-length=30 [sample.bam] cuffmerge -s [refer.genome.fa] -g [gene.gff] [sample.gtf.list]
- Genetic Marker
View Sequences
All of these feature-box tracks are clickable:
- Gene Locus and Principle Transcript tracks: Clicking them would go to relevant gene profile or transcript profile page.
- Other tracks: Clicking them would show a pop-up box displaying the region sequences
Save Track Data
In this track, data would be saved as GFF/BED format text of the current region
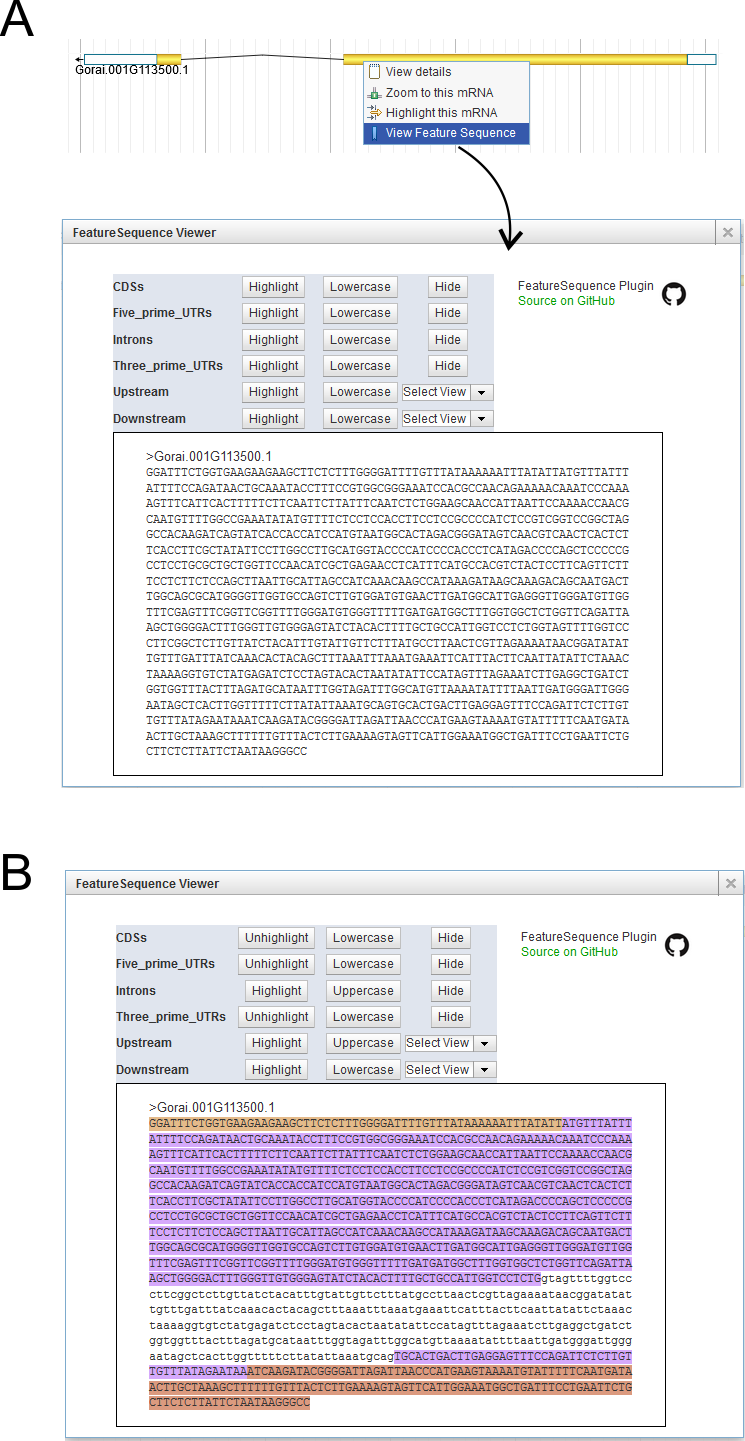
Transcript Feature
For the Principle Transcript track, CottonFGD applies the JBrowse FeatureSequence plugin (Fig. 9A). It is very easy to color and retrieve exon, intron, UTR, CDS, ... sequences. Including:
- Highlight feature elements (color is random)
- Toggle lowercase/Uppercase
- Hide/Show certain features. For example, you can hide introns to get spliced mRNA sequences
Fig. 9B shows an example. The UTR sequences are marked in brown, introns in lowercase and CDSs in purple. This colored sequence can be directly copied and pasted to other rich-text software (such as MS-Word).
Track Type: XYPlot
These tracks display quantitative data (Y axis) distribution among the reference genome (X axis). Hovering on it would show y values (Fig. 4C1, RNA-seq Coverage and C2, GC Content).