|
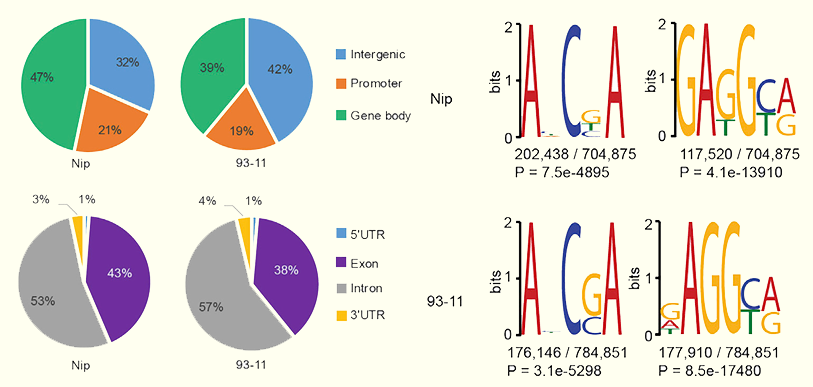
DNA N6-methyladenosine (6mA), a new epigenetic mark has been identified as a major breakthrough and widely distributed throughout the eukaryote genomes, which is less abundant and more sensitive compared to 5mC. Analysis of 6mA genomic distribution and biological function showed that 6mA is associated with gene expression, plant development and stress responses, suggesting that 6mA hallmarked an important and crucial role in rice. In this part, 6mA and 5mC methylation information has been refined to permit searching by selecting a candidate chromosome and entering the site of interest (from the starting site to the ending site); it then links to detailed pages. For 6mA, we provide detailed methylation parameters (e.g., chromosome position, DNA strand, and fraction score), and a 20-bp reference sequence above and below the given methylation site is proffered. In contrast, for 5mC methylation, we have provided methylation types (CG, CHG and CHH) but not detailed methylation parameters. Moreover, genomic sequences at a given methylation site (i.e., whether the methylation site is within a gene or in an intergenic region) are available as well.
|
|