Overview
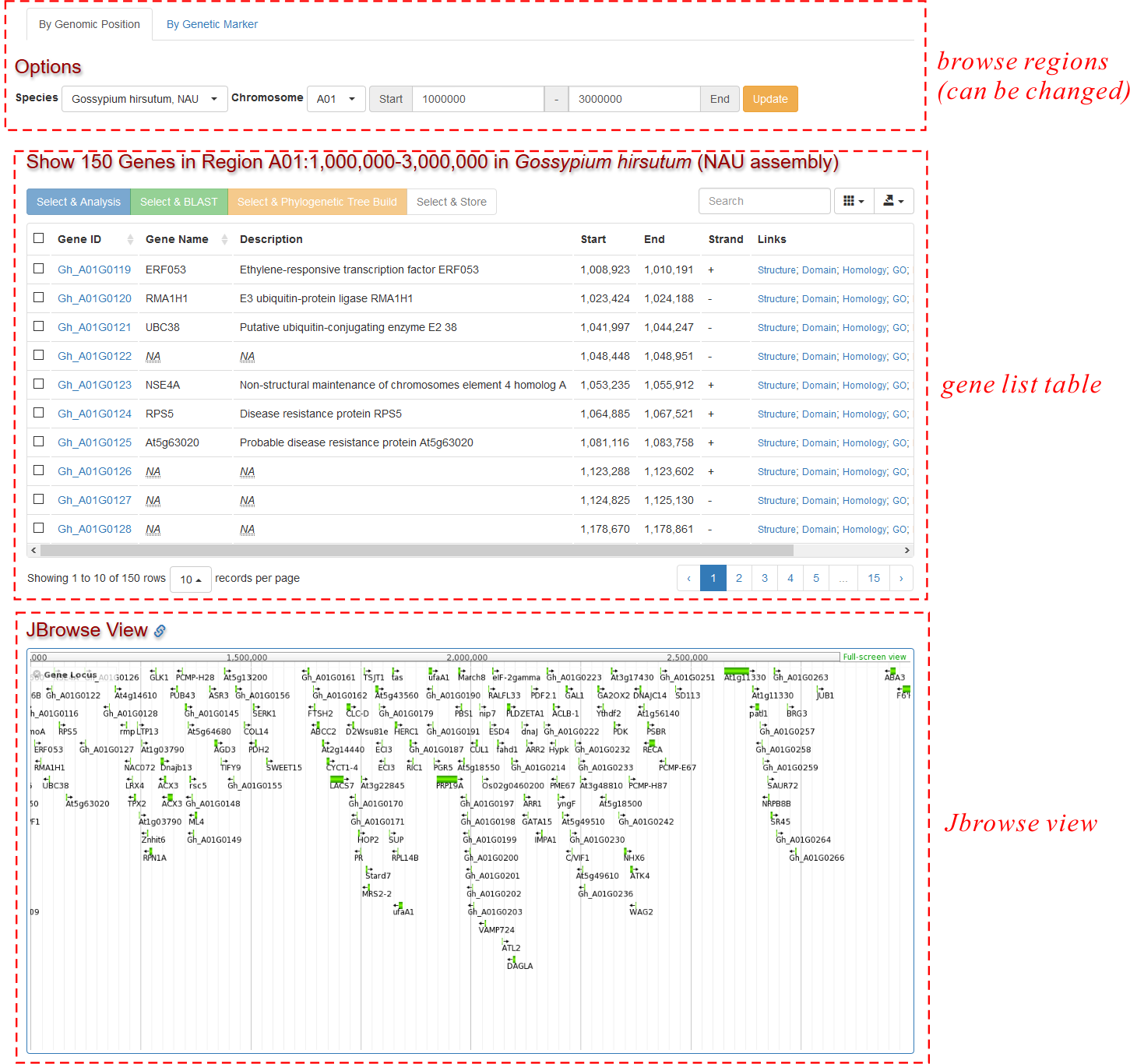
This page consists of three parts (See Fig. 1):
- A region table showing the current chromosomal region. The default region is 1,000,000-2,000,000 in chromosome A01 in Gossypium hirsutum (CRI) (with 100 genes).It can be changed by users, and the browsers would "remember" users' last change.
- A gene list table showing all the annotated genes in the selected chromosomal region, including some basic information: gene ID, name, description, genomic coordinates and some links to the transcript profile page.
- An embedded JBrowse view of the current chromosomal region. Here users can see the distribution of genes in the current chromosomal region in a more visualized way.
Change Your Region
By Genomic Position
Users can change the species, chromosome, start and end to any regions they want. After clicking the Update button, the page would be refreshed to show genes on newly selected regions. Note: The "Start" and "End" field only accept integer numbers larger than 1.
By Genetic Marker
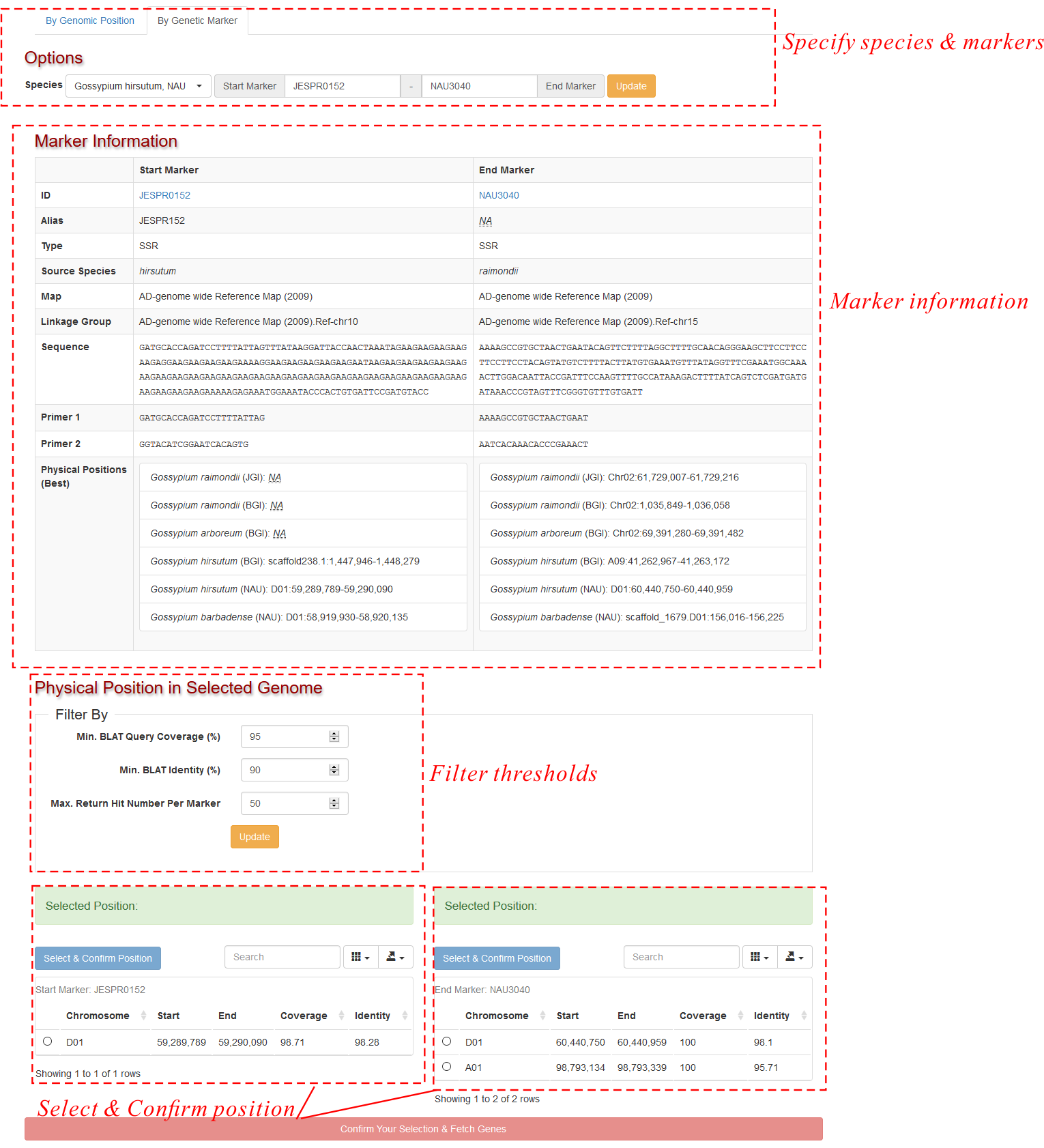
If you want to browse by genetic marker, switch to the "By Genetic Marker" Tab. Specify your target species and a pair of genetic marker IDs (SSR, RFLP or InDel). The default value is a pair of SSR marker: NBRI_GE19612 and HAU2217. After clicking the update button, CottonFGD would show some basic information of these two markers (imported from CottonGen) and the possible mapping sites in the reference genome (Fig. 2). The mapping sites are ordered by their mapping scores.
Users should select the sites they want (usually the top is better) by clicking the first-column radios and confirm their selection by clicking the top-left confirm button. Note: The selected two sites must be on the same chromosome. Finally, after clicking the buttom button, CottonFGD displays the result gene list table similar as Fig. 1.
View in JBrowse
Below the gene list table, a snapshot of JBrowse view of the current chromosomal region is shown. Users can also click the attached link to jump to the new JBrowse window.